Command line tools#
In addition to the Python objects presented in previous sections, pyALF offers a set of scripts that make it easy to leverage pyALF from a Unix shell (e.g. Bash or zsh). They are located in the folder py_alf/cli and, as mentioned in Prerequisites and installation, it is recommended to add this folder to the $PATH environment variable, to conveniently use the scripts: \(\phantom{\xi}\)
export PATH="/path/to/pyALF/py_alf/cli:$PATH"
The list of all command line tools can be found in the reference. Out of those, this section will only introduce two more elaborate scripts, namely alf_run.py and alf_postprocess.py.
When starting a code line in Jupyter with an exclamation mark, the line will be interpreted as a shell command. We will use this feature to demonstrate the shell tools. Through this,
alf_run.py#
The script alf_run.py enables most of the features displayed in Compiling and running ALF to be used directly from the shell. The help text lists all possible arguments:
!alf_run.py -h
usage: alf_run.py [-h] [--alfdir ALFDIR] [--sims_file SIMS_FILE]
[--branch BRANCH] [--machine MACHINE] [--mpi]
[--n_mpi N_MPI] [--mpiexec MPIEXEC]
[--mpiexec_args MPIEXEC_ARGS] [--do_analysis]
Helper script for compiling and running ALF.
optional arguments:
-h, --help show this help message and exit
--alfdir ALFDIR Path to ALF directory. (default: os.getenv('ALF_DIR',
'./ALF')
--sims_file SIMS_FILE
File defining simulations parameters. Each line starts
with the Hamiltonian name and a comma, after wich
follows a dict in JSON format for the parameters. A
line that says stop can be used to interrupt.
(default: './Sims')
--branch BRANCH Git branch to checkout.
--machine MACHINE Machine configuration (default: 'GNU')
--mpi mpi run
--n_mpi N_MPI number of mpi processes (default: 4)
--mpiexec MPIEXEC Command used for starting a MPI run (default:
'mpiexec')
--mpiexec_args MPIEXEC_ARGS
Additional arguments to MPI executable.
--do_analysis, --ana Run default analysis after each simulation.
For example, to run a series of four different simulations of the Kondo model, the first step is to create a file specifying the parameters, with one line per simulation:
!cat Sims_Kondo
Kondo, {"L1": 4, "L2": 4, "Ham_JK": 0.5}
Kondo, {"L1": 4, "L2": 4, "Ham_JK": 1.0}
Kondo, {"L1": 4, "L2": 4, "Ham_JK": 1.5}
Kondo, {"L1": 4, "L2": 4, "Ham_JK": 2.0}
Then, one can execute alf_run.py with options as desired, the script automatically recompiles ALF for each simulation. For understanding some of the options, the section Compiling and running ALF might help. The following printout is truncated for brevity.
!alf_run.py --sims_file ./Sims_Kondo --mpi --n_mpi 4 --mpiexec orterun
Number of simulations: 4
Compiling ALF...
Cleaning up Prog/
Cleaning up Libraries/
Cleaning up Analysis/
Compiling Libraries
ar: creating modules_90.a
ar: creating libqrref.a
Compiling Analysis
Compiling Program
Parsing Hamiltonian parameters
filename: Hamiltonians/Hamiltonian_Kondo_smod.F90
filename: Hamiltonians/Hamiltonian_Hubbard_smod.F90
filename: Hamiltonians/Hamiltonian_Hubbard_Plain_Vanilla_smod.F90
filename: Hamiltonians/Hamiltonian_tV_smod.F90
filename: Hamiltonians/Hamiltonian_LRC_smod.F90
filename: Hamiltonians/Hamiltonian_Z2_Matter_smod.F90
Compiling program modules
Link program
Done.
Prepare directory "/scratch/pyalf-docu/doc/source/usage/ALF_data/Kondo_L1=4_L2=4_JK=0.5" for Monte Carlo run.
Create new directory.
Run /home/jschwab/Programs/ALF/Prog/ALF.out
ALF Copyright (C) 2016 - 2021 The ALF project contributors
This Program comes with ABSOLUTELY NO WARRANTY; for details see license.GPL
This is free software, and you are welcome to redistribute it under certain conditions.
No initial configuration
Compiling ALF...
...
...
...
alf_postprocess.py#
The script alf_postprocess.py enables most of the features discussed in Postprocessing, except for plotting capabilities, to be used directly from the shell. The help text lists all possible arguments:
!alf_postprocess.py -h
usage: alf_postprocess.py [-h] [--check_warmup] [--check_rebin]
[-l CHECK_LIST [CHECK_LIST ...]] [--do_analysis]
[--always] [--gather] [--no_tau]
[--custom_obs CUSTOM_OBS] [--symmetry SYMMETRY]
[directories ...]
Script for postprocessing Monte Carlo bins.
positional arguments:
directories Directories to analyze. If empty, analyzes all
directories containing file "data.h5" it can find.
optional arguments:
-h, --help show this help message and exit
--check_warmup, --warmup
Check warmup.
--check_rebin, --rebin
Check rebinning for controlling autocorrelation.
-l CHECK_LIST [CHECK_LIST ...], --check_list CHECK_LIST [CHECK_LIST ...]
List of observables to check for warmup and rebinning.
--do_analysis, --ana Do analysis.
--always Do not skip analysis if parameters and bins are older
than results.
--gather Gather all analysis results in one file named
"gathered.pkl", representing a pickled pandas
DataFrame.
--no_tau Skip time displaced correlations.
--custom_obs CUSTOM_OBS
File that defines custom observables. This file has to
define the object custom_obs, needed by
py_alf.analysis. (default: os.getenv("ALF_CUSTOM_OBS",
None))
--symmetry SYMMETRY, --sym SYMMETRY
File that defines lattice symmetries. This file has to
define the object symmetry, needed by py_alf.analysis.
(default: None))
To use the symmetrization feature, one needs a file defining the object symmetry, similar to the already used file custom_obs.py defining custom_obs.
!cat sym_c4.py
"""Define C_4 symmetry (=fourfold rotation) for pyALF analysis."""
from math import pi
# Define list of transformations (Lattice, i) -> new_i
# Default analysis will average over all listed elements
def sym_c4_0(latt, i): return i
def sym_c4_1(latt, i): return latt.rotate(i, pi*0.5)
def sym_c4_2(latt, i): return latt.rotate(i, pi)
def sym_c4_3(latt, i): return latt.rotate(i, pi*1.5)
symmetry = [sym_c4_0, sym_c4_1, sym_c4_2, sym_c4_3]
To analyze the results from the Kondo model and gather them all in one file gathered.pkl, we execute the following command. The printout has again been truncated.
!alf_postprocess.py --custom_obs custom_obs.py --symmetry sym_c4.py --ana --gather ALF_data/Kondo*
### Analyzing ALF_data/Kondo_L1=4_L2=4_JK=0.5 ###
/scratch/pyalf-docu/doc/source/usage
Custom observables:
custom E_squared ['Ener_scal']
custom E_pot_kin ['Pot_scal', 'Kin_scal']
custom SpinZ_pipi ['SpinZ_eq']
Scalar observables:
Constraint_scal
Ener_scal
Kin_scal
Part_scal
Pot_scal
Histogram observables:
Equal time observables:
Den_eq
Dimer_eq
Green_eq
SpinZ_eq
Time displaced observables:
Den_tau
Dimer_tau
Green_tau
Greenf_tau
SpinZ_tau
### Analyzing ALF_data/Kondo_L1=4_L2=4_JK=1.0 ###
/scratch/pyalf-docu/doc/source/usage
...
...
...
ALF_data/Kondo_L1=4_L2=4_JK=0.5
ALF_data/Kondo_L1=4_L2=4_JK=1.0
ALF_data/Kondo_L1=4_L2=4_JK=1.5
ALF_data/Kondo_L1=4_L2=4_JK=2.0
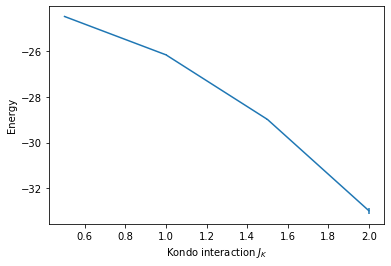
The data from gathered.pkl can, for example, be read and plotted like this:
# Import modules
import pandas as pd
import matplotlib.pyplot as plt
# Load pickled DataFrame
res = pd.read_pickle('gathered.pkl')
# Create figure with axis labels
fig, ax = plt.subplots()
ax.set_xlabel(r'Kondo interaction $J_K$')
ax.set_ylabel(r'Energy')
# Plot data
ax.errorbar(res.ham_jk, res.Ener_scal0, res.Ener_scal0_err);